Congrats to Castoe lab PhD students Aundrea Westfall, Sid Gopalan and other coauthors for their recent paper in Genome Biology and Evolution. This paper provides exciting new evidence that the evolution of new gene regulatory networks may involve the recruitment of existing pathways that are inherently heterogeneous, and this may be a strategy for avoiding particular cellular constraints. This study used snake venom glands and venom systems as an empirical platform for exploring and testing this concept.
Westfall, A.K.*, S.S. Gopalan*, B.W. Perry, R.H. Adams, A. Saviola, S.P. Mackessy, and T.A. Castoe. 2023. Single-cell heterogeneity in snake venom expression is hardwired by co-option of regulators from progressively activated pathways. Genome Biology and Evolution 15: evad109. *joint primary authors. PDF Supplementary Data
Understanding how new complex physiological traits arise via the evolution of gene regulatory networks (GRNs), and how cellular variation in GRNs manifest in organismal phenotypes, are fundamental to understanding the processes that generate phenotypic diversity. A growing appreciation that cellular heterogeneity underlies many organism-level phenotypes raises new questions about what factors drive this heterogeneity and how complex heterogeneous systems and the GRNs that control them evolve.
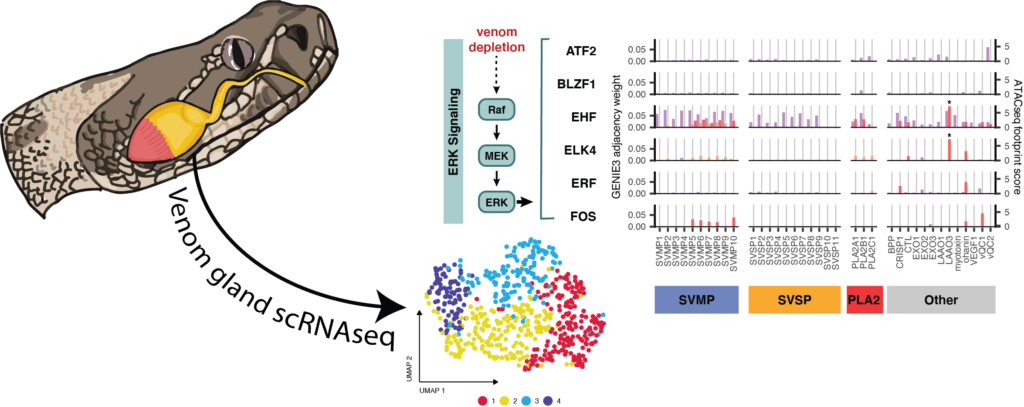
One emerging model to study the evolution of novel complex traits – and the evolutionary co-option of genes and regulatory systems that underlie them – are snake venom systems, including the specialized secretory glands that produce venom. Snake venoms are comprised of complex mixtures of toxic proteins expressed in specialized secretory glands that exhibit spatial heterogeneity in venom toxin expression. Prior studies have hypothesized that cellular constraints might drive this heterogeneity, while others have separately implicated the co-option of suites of conserved GRNs to regulate venom production.
Here, we integrate single-nucleus RNA sequencing of a rattlesnake venom gland, along with existing tissue-level chromatin accessibility data, to explore the relationships between cellular heterogeneity and GRNs underlying venom gene expression. We find evidence that venom gene expression is highly heterogeneous across otherwise indistinguishable secretory cells, and that this variation correlates with activation of key pathways proposed to regulate venom. We further demonstrate that snake venom genes have recruited transcription factors from existing GRNs that exhibit inherently heterogeneous activity in secretory cells, including distinct combinations of ERK and unfolded-protein response (UPR) pathway transcription factors that regulate suites of venom genes via the phased activation of these pathways.
These findings broadly suggest that there may be more complex constraints on the evolution of GRNs than previously appreciated – including having to comply with constraints imposed by cellular physiology and chromatin. Our inferences highlight an insightful evolutionary solution to this problem in snake venom, where new gene regulatory network components appear to have been co-opted from an established highly dynamic set of regulatory cascades (ERK and UPR) that exhibit phased responses in secretory cells. This strategy effectively generates expression heterogeneity within and among venom gene families, suggesting that similar constraints and analogous evolutionary solutions may be relevant for the re-wiring of GRNs in other multi-gene families expressed in the same tissue. Collectively, this and prior studies suggest that these two conserved pathways may be particularly relevant raw material for co-option by new GRNs in which extremely high expression of multiple gene families is the outcome favored by selection.
.