Congrats to PhD student Sid Gopalan, and lab alum Blair and Drew, on a new paper that identifies the evolutionary origins, genomic structure and copy number variation, and regulatory architecture of snake venom myotoxins.
Gopalan, S.S., B.W. Perry, D.R. Schield, C. Smith, S.P. Mackessy, and T.A. Castoe. 2022. Origins, genomic structure, and copy number variation of snake venom myotoxins. Toxicon PDF
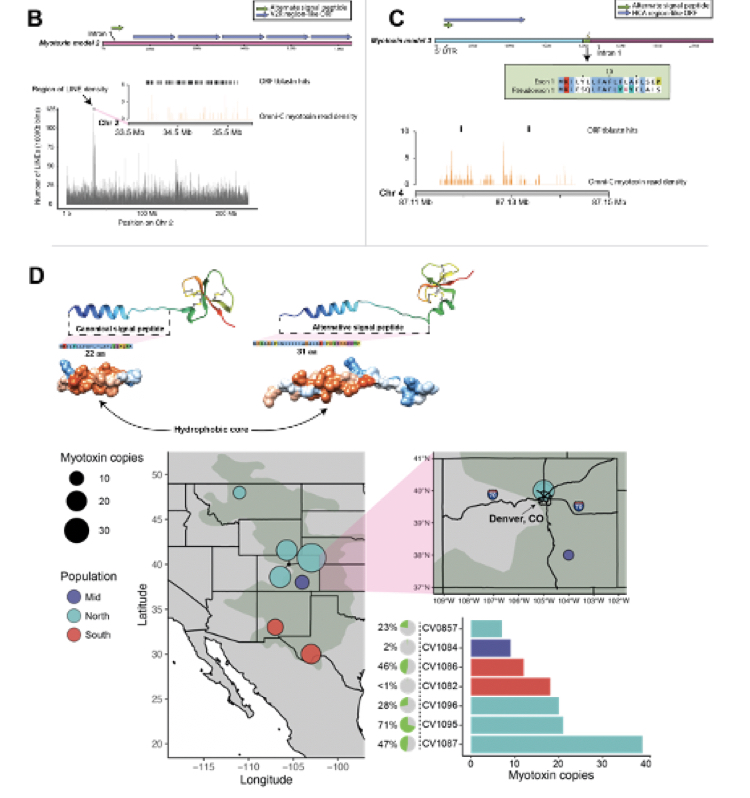
Crotamine, myotoxin a and homologs are short peptides that often comprise major fractions of rattlesnake venoms and have been extensively studied for their bioactive properties. These toxins are thought to be important for rapidly immobilizing mammalian prey and are implicated in serious, and sometimes fatal, responses to envenomation in humans. While high quality reference genomes for multiple venomous snakes are available, the loci that encode myotoxins have not been successfully assembled in any existing genome assembly. Here, we integrate new and existing genomic and transcriptomic data from the Prairie Rattlesnake (Crotalus viridis viridis) to reconstruct, characterize, and infer the chromosomal locations of myotoxin-encoding loci. We integrate long-read mRNA (Pacific Bioscience’s Iso-Seq) and short-read RNA-seq to infer gene sequence diversity and characterize patterns of myotoxin and paralogous b-defensin expression across multiple tissues. We also identify two long non-coding RNA sequences which both encode functional myotoxins, demonstrating a newly discovered source of venom coding sequence diversity. We also integrate long-range mate-pair chromatin contact data and linked-read sequencing to infer the structure and chromosomal locations of the three myotoxin-like loci. Further, we conclude that the venom-associated myotoxin is located on chromosome 1 and is adjacent to non-venom paralogs. Consistent with this locus contributing to venom composition, we find evidence that the promoter of this gene is selectively open in venom gland tissue and contains transcription factor binding sites implicated in broad trans-regulatory pathways that regulate snake venoms. This study provides the best genomic reconstruction of myotoxin loci to date and raises questions about the physiological roles and interplay between myotoxin and related genes, as well as the genomic origins of snake venom variation.